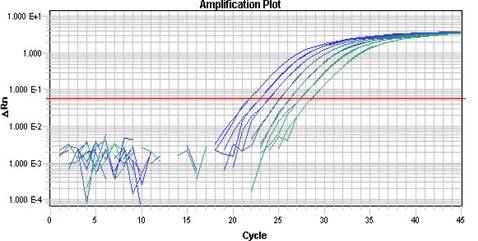
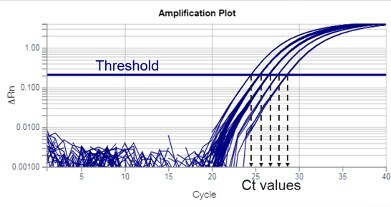
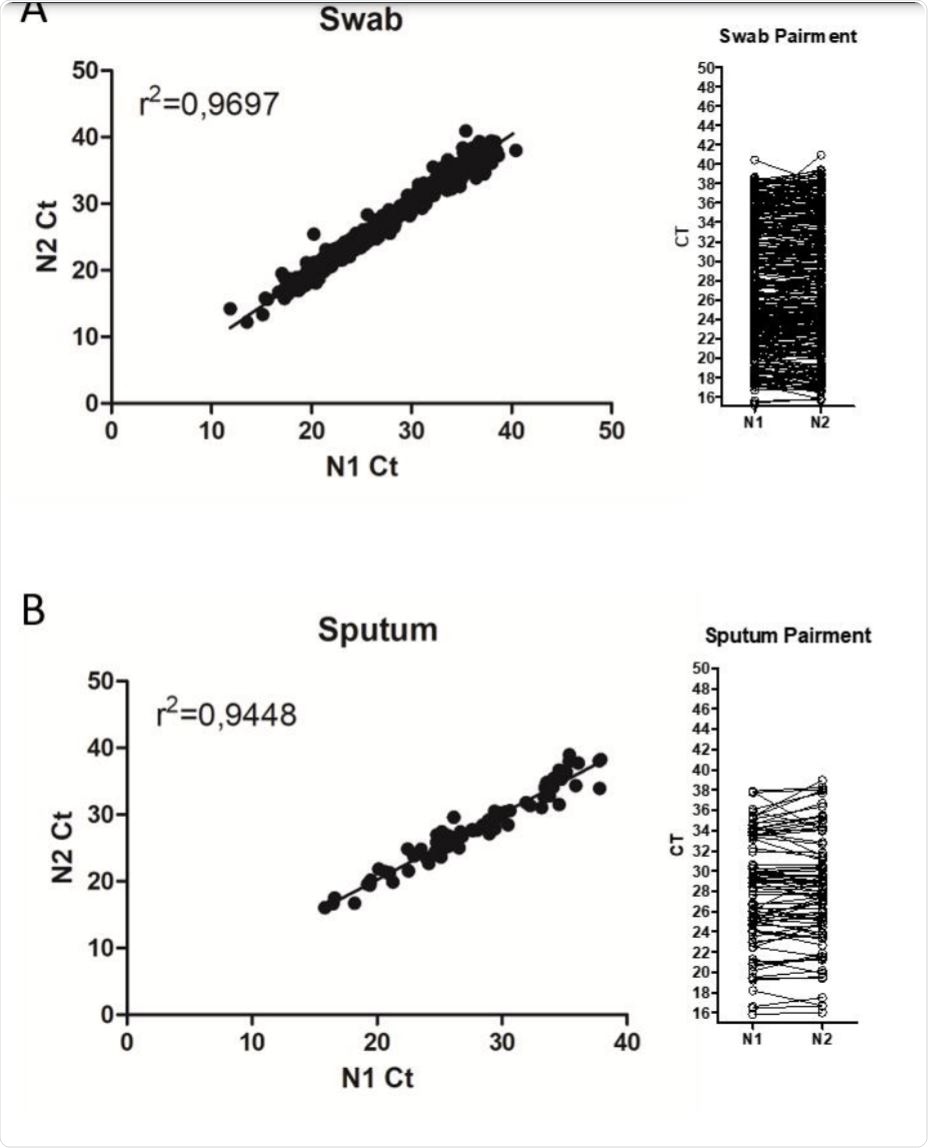
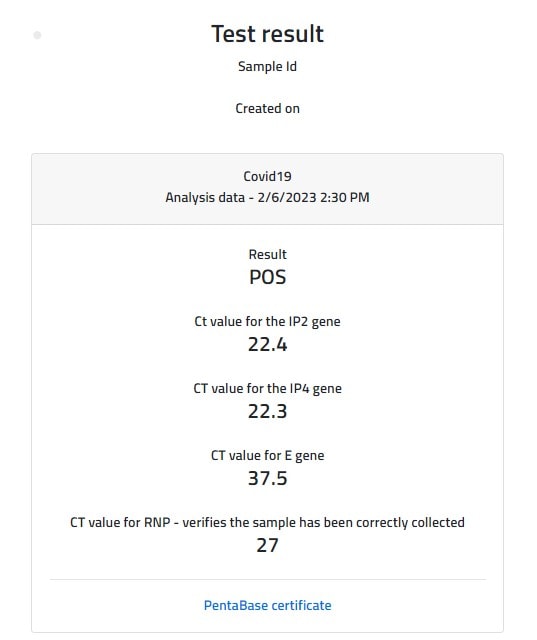
Variability of cycle threshold values in an external quality assessment scheme for detection of the SARS-CoV-2 virus genome by RT-PCR
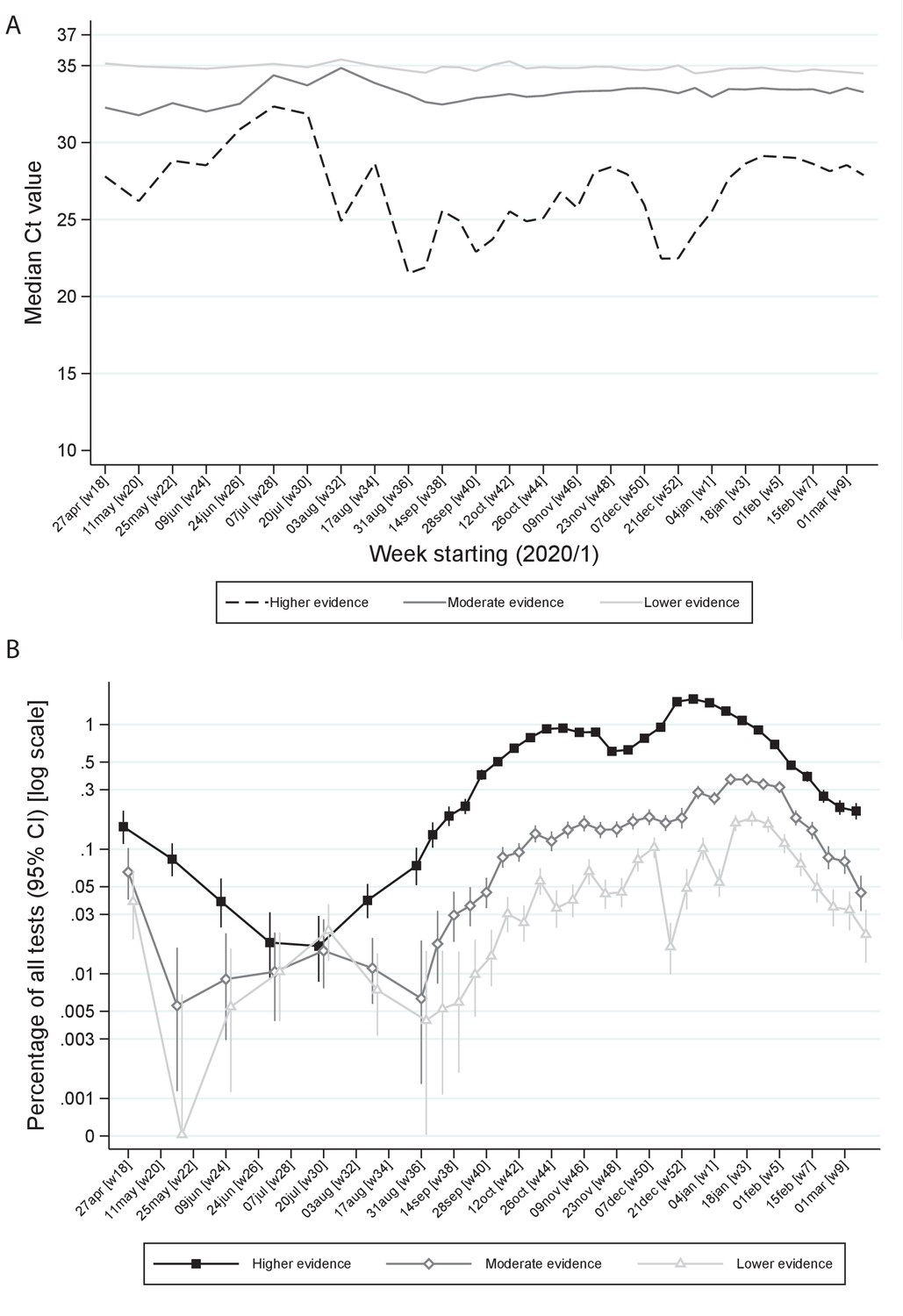
Ct threshold values, a proxy for viral load in community SARS-CoV-2 cases, demonstrate wide variation across populations and over time | eLife

Change in ct value of RNA positive sample with low ct value (high viral... | Download Scientific Diagram

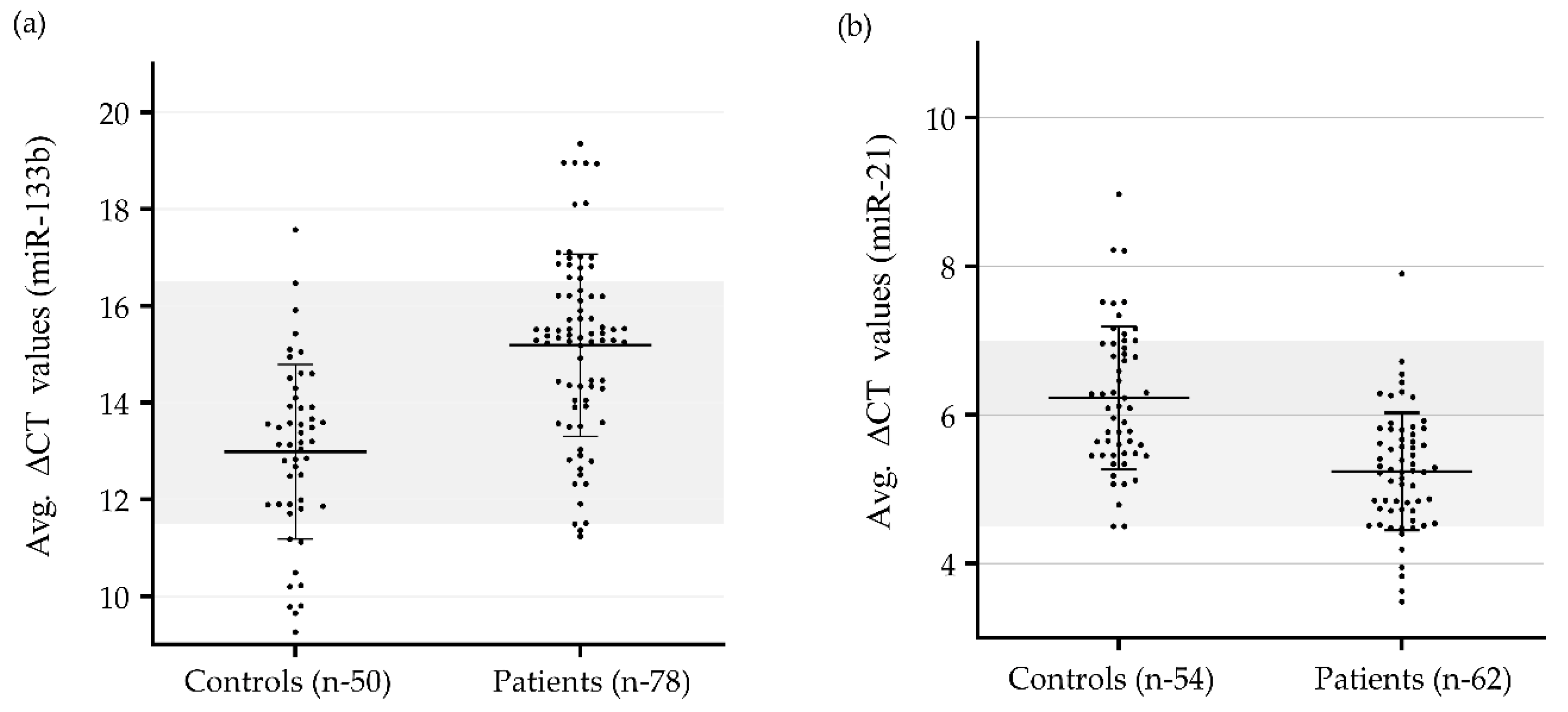
Choice of endogenous control for gene expression in nonsmall cell lung cancer | European Respiratory Society

Improved RT-PCR SARS-Cov2 results interpretation by indirect determination of cut-off cycle threshold value | medRxiv

Ct threshold values, a proxy for viral load in community SARS-CoV-2 cases, demonstrate wide variation across populations and over time | eLife

Cycle threshold values in RT‐PCR to determine dynamics of SARS‐CoV‐2 viral load: An approach to reduce the isolation period for COVID‐19 patients - Aranha - 2021 - Journal of Medical Virology -
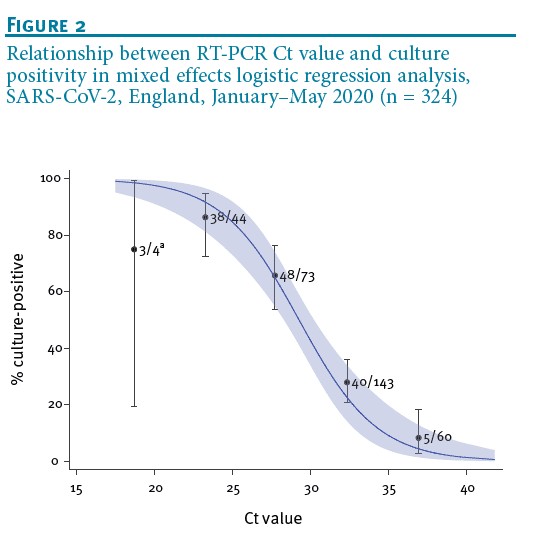
Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19 in England. - The Centre for Evidence-Based Medicine

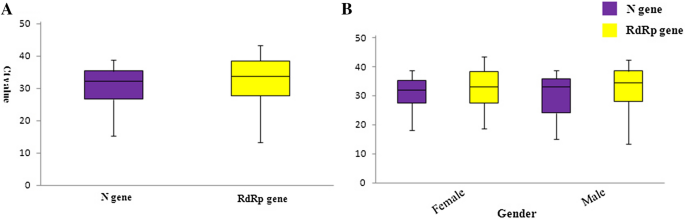
SARS-CoV-2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a widely used commercial multiplex PCR assay | medRxiv

SARS-CoV-2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a commercial multiplex PCR assay - ScienceDirect
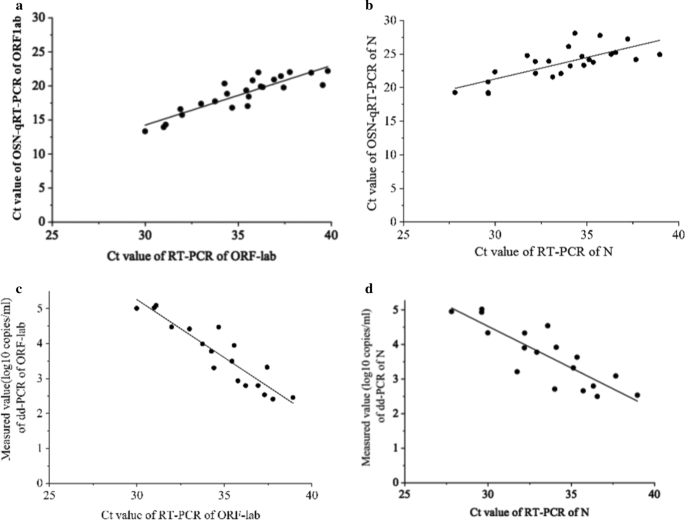
Analysis and validation of a highly sensitive one-step nested quantitative real-time polymerase chain reaction assay for specific detection of severe acute respiratory syndrome coronavirus 2 | Virology Journal | Full Text
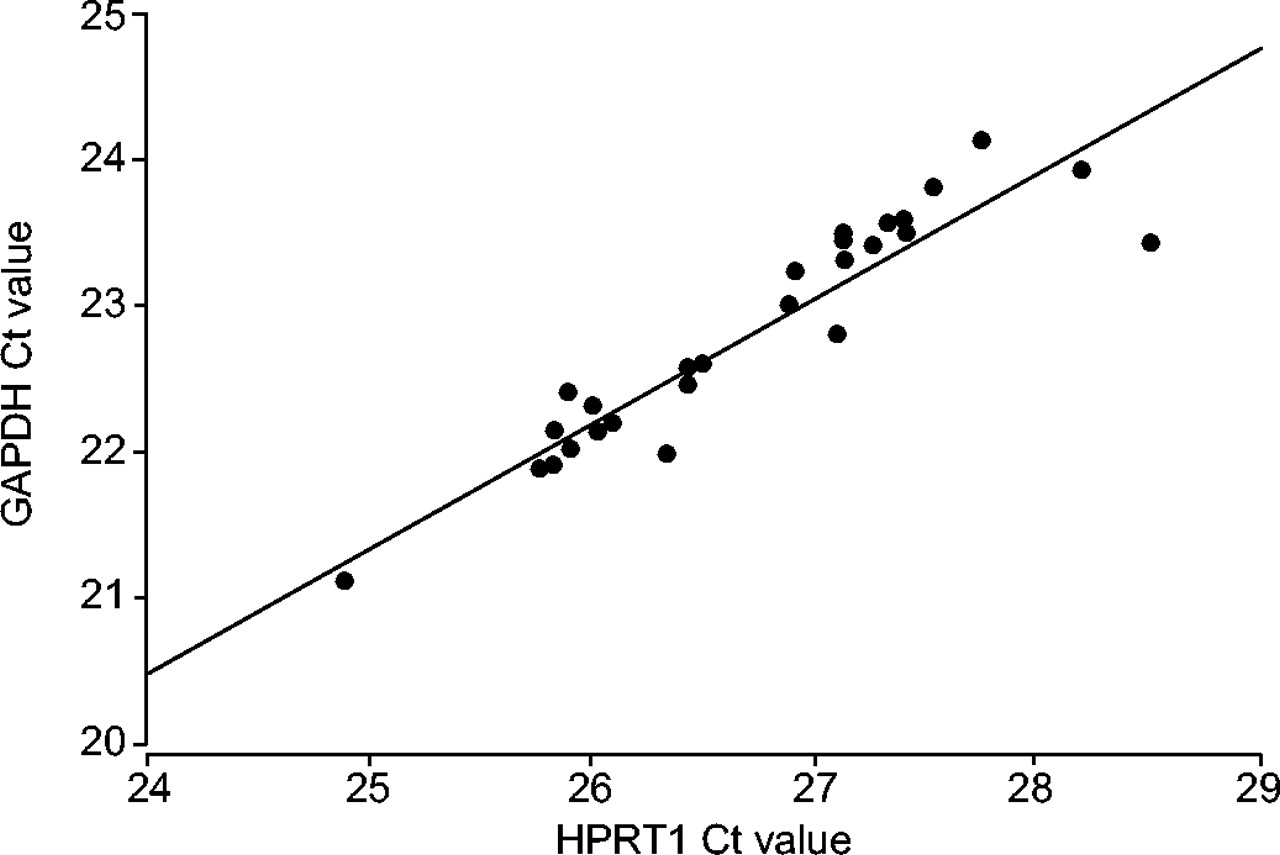
Choice of endogenous control for gene expression in nonsmall cell lung cancer | European Respiratory Society

Pooling of samples for testing for SARS-CoV-2 in asymptomatic people - The Lancet Infectious Diseases
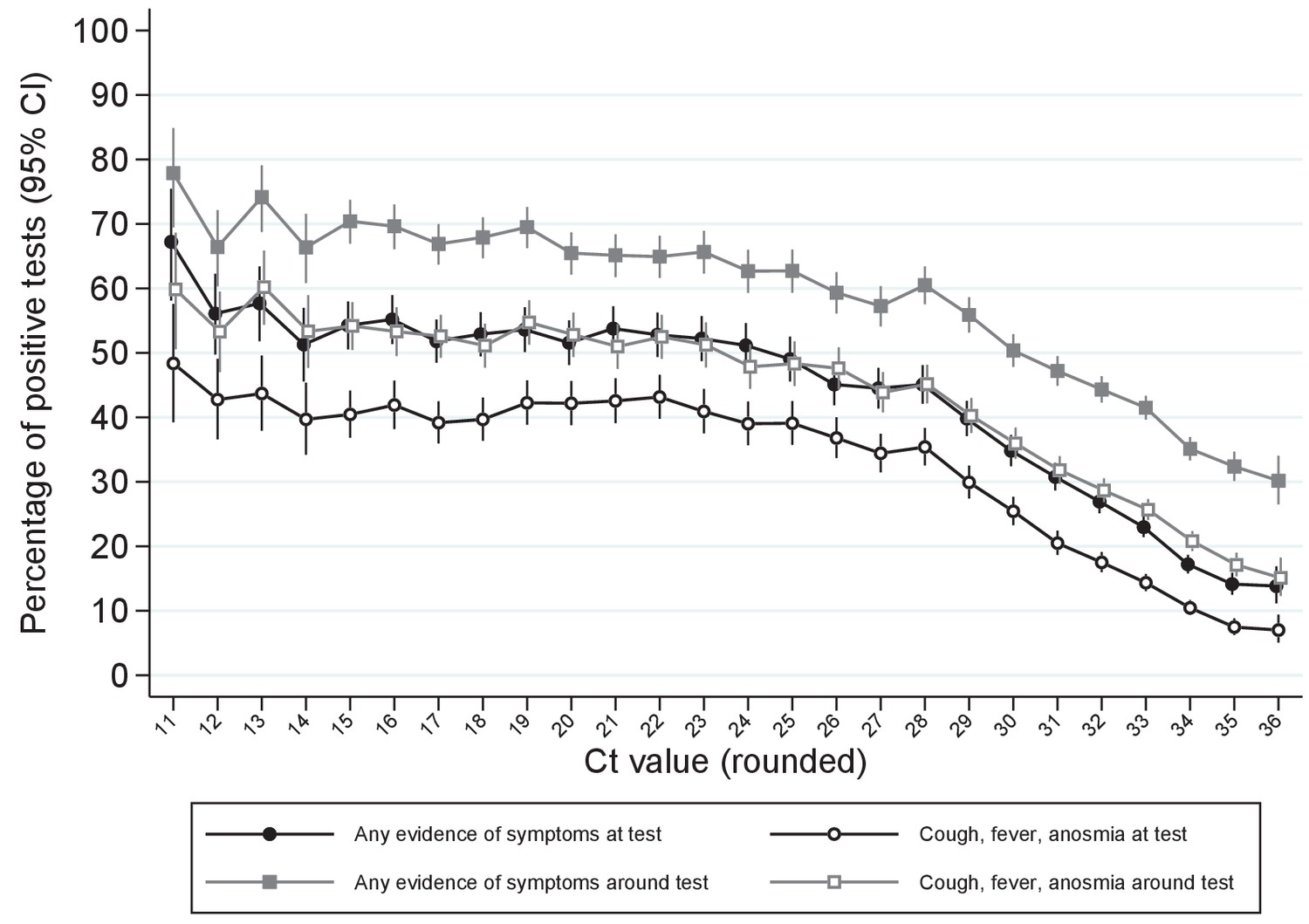
Ct threshold values, a proxy for viral load in community SARS-CoV-2 cases, demonstrate wide variation across populations and over time | eLife
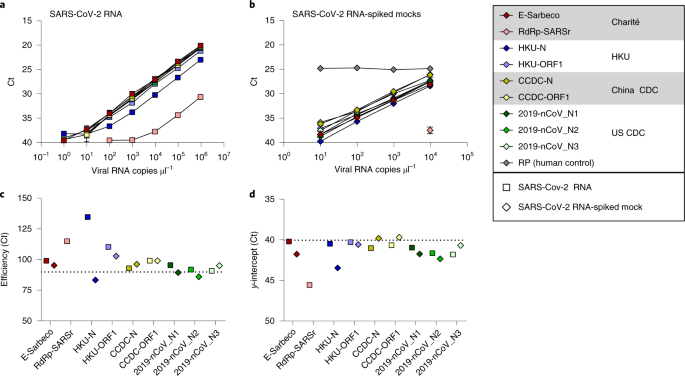
Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets | Nature Microbiology